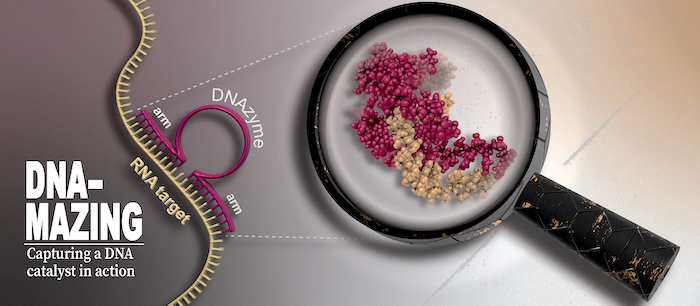
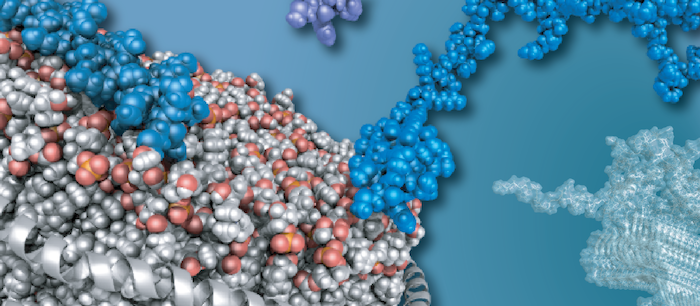
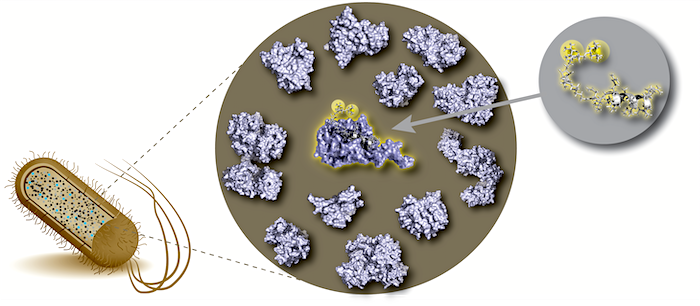
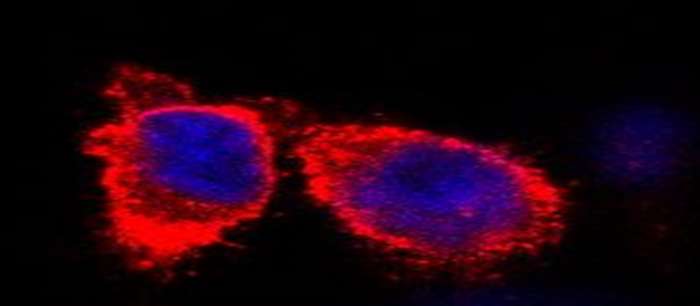
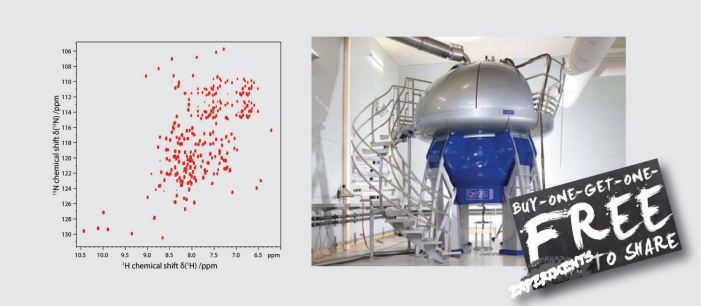
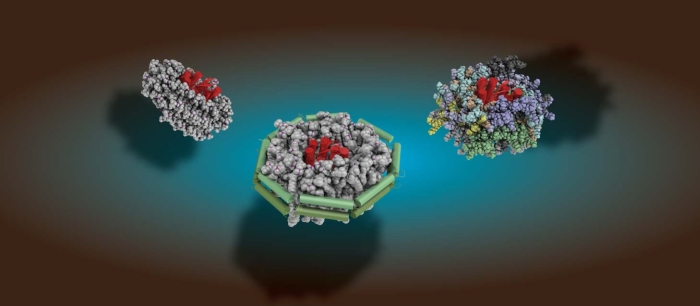







To understand life on a molecular level it is of fundamental importance to study structure and function of life's key players -proteins, nucleic acids and lipids - in their native environment.
Our main focus is to develop and apply tailored methods to overcome current limitations in structural biology and to obtain high-resolution insights into life's key players in an increasingly native setting. learn more about our research -here-
We are very gratefull to support from: